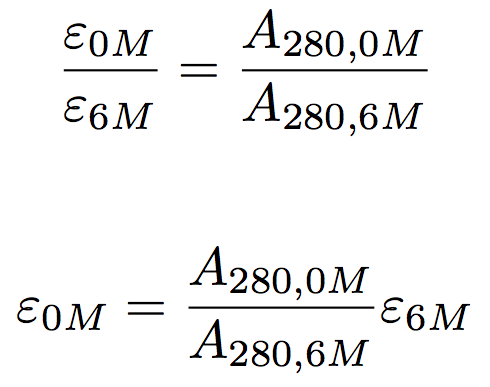How To Calculate Molar Extinction Coefficient Of A Protein

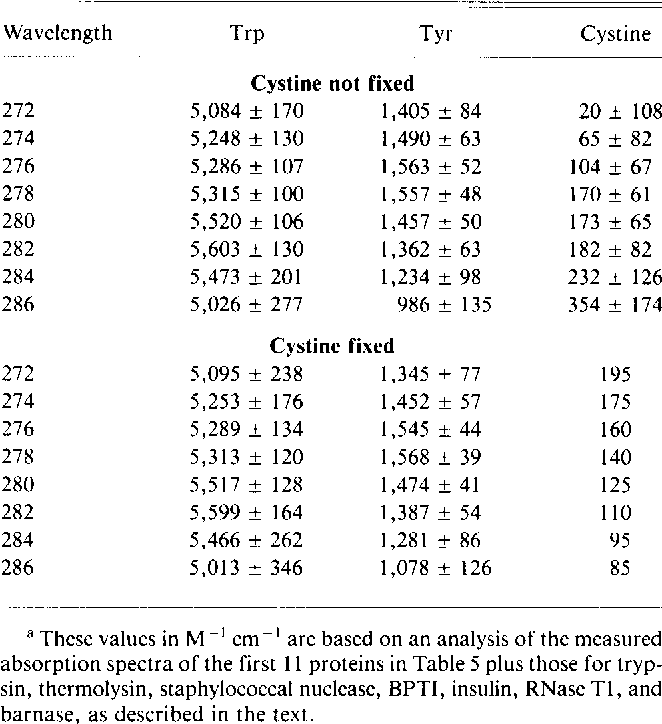
Table 2 From Calculation Of Protein Extinction Coefficients From Amino Acid Sequence Data Semantic Scholar
www.semanticscholar.org
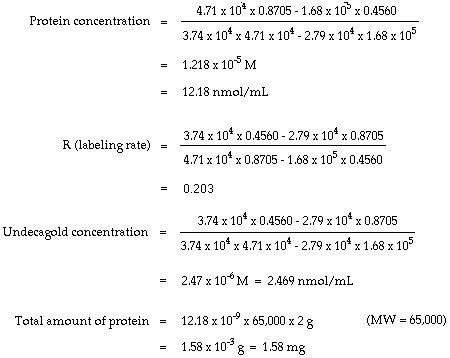
Epsilon 280 m 1 cm 1 trp 5500 tyr 1490 cystine 125 these epsilon 280 values are quite reliable for proteins containing trp residues and less reliable for proteins that do not.

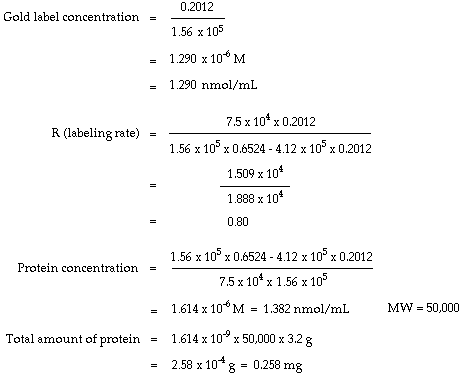
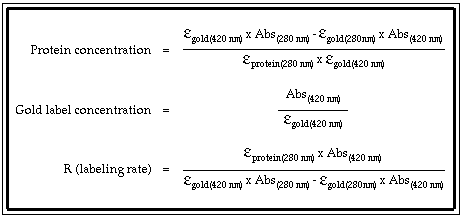
How to calculate molar extinction coefficient of a protein. Emgdn hclaemtyr bemtrp cemcys. Enter the absorbance and molar concentration into the calculator to determine the molar extinction coefficient. An extinction coefficient can also be predicted by a theoretical calculation from the number of a280 absorbing residues trp tyr cystine disulfide bonds.
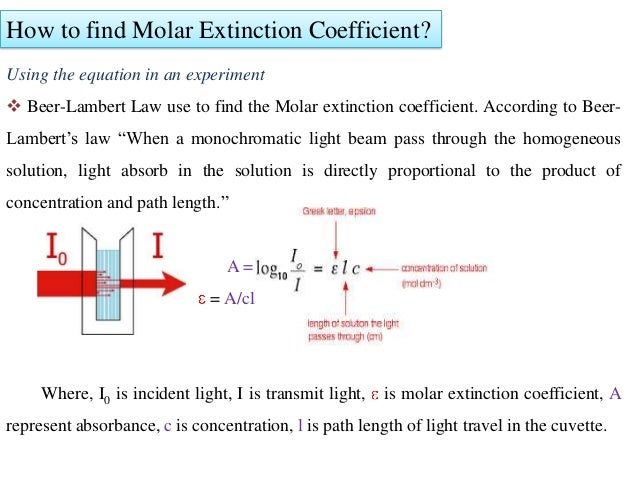
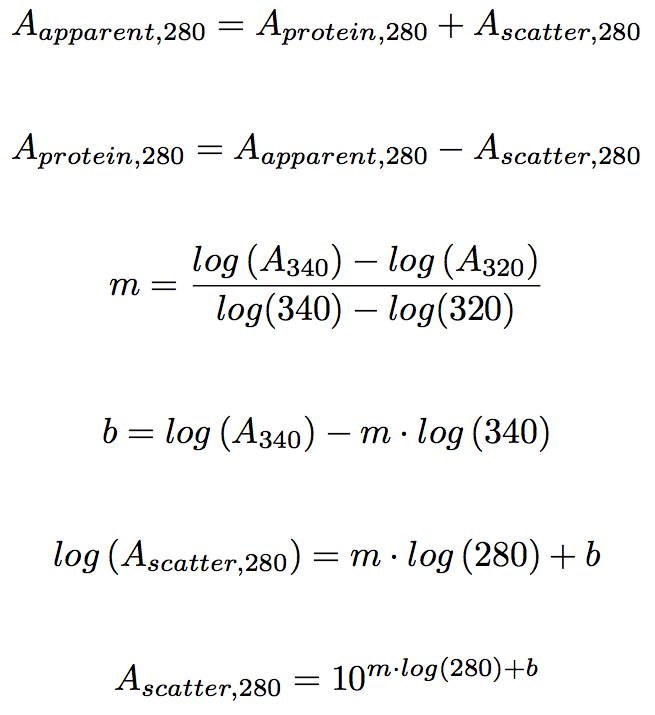
Where abc are the number of tyrosine trytophan and cystine residues per mole of protein and e residue are the molar extinction rated of the residue at the wavelength used 280 nm. Based on a sample of 116 measured epsilon values for 80 proteins the epsilon at 280 nm of a folded protein in water epsilon 280 can best be predicted with this equation. If you measure the absorbance of two samples one in native and one in denaturing conditions like 6 m guanidine hydrochloride with exactly the same protein amount you can calculate the.
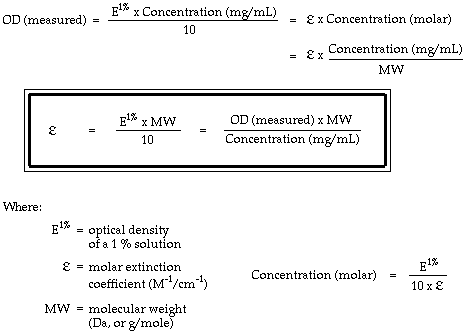
The relationship between molar extinction coefficient e molar and percent extinction coefficient e percent is as follows. Total of residues. Numbers of individual residues.
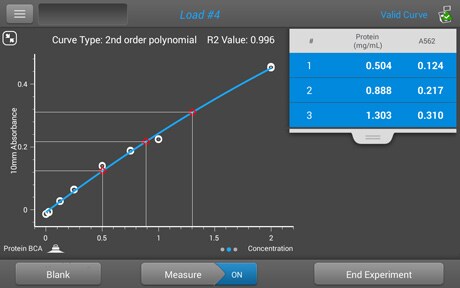
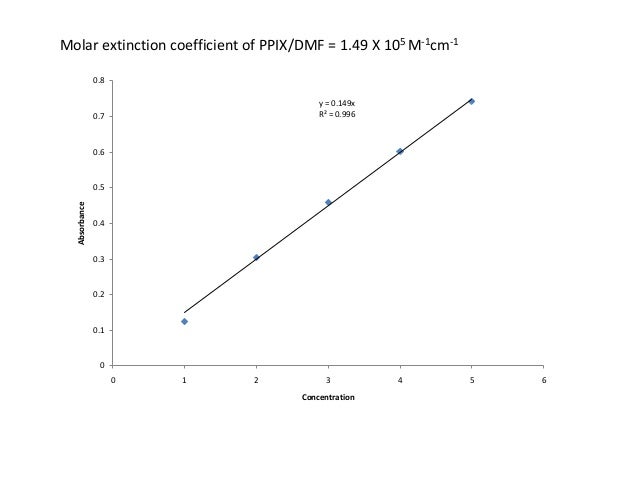
Protein extinction coefficient calculator. It is possible to determine the extinction coefficient of a protein experimentally. To get the extinction coefficient of the native protein beers law is used.
Enter your peptide sequence below using single letter code. You do this by a280 measurements of a dilution series of the protein in known concentrations. Where 280 is the molar extinction coefficient at 280 nm and n is the number of corresponding residues present in the protein.
Ala a asx b cys c asp d. 280 nw x 5500 ny x 1490 nc x 125. If the number of absorbing side chains in the amino acid sequence of a protein is known the specific extinction coefficient at 280 nm can be estimated using the following formula.

User Nader A Khamis Notebook Experimental Biological Chemistry Au Fall 2011 2011 09 14 Openwetware
openwetware.org






















/beers-law-definition-and-equation-608172_FINAL-20ddc4fef437472db0a0ebe395770c76.png)